Are Double Strand Repairs On Mcat
Achieve Mastery of Medical Concepts
Table of Contents
Overview of DNA Harm and Repair
Etiology
Dna damage tin be acquired by:
- Environmental exposures:
- Sunlight/UV radiation
- Radiation
- Chemicals
- Viruses
- Diet
- Reactive oxygen species (ROS) produced during normal metabolic processes
- Replication errors
- Age
Types of Deoxyribonucleic acid damage
- Single-stranded damage:
- Damage to 1 strand allows use of the other strand every bit a template for repair.
- Examples of single-stranded harm include:
- Breaks in 1 strand of the Dna
- Mismatched base pairs
- Incorporation of uracil into the Deoxyribonucleic acid
- Oxidation or alkylation of the DNA
- Double-stranded breaks:
- Both strands of Deoxyribonucleic acid are severed.
- Dangerous to genome stability (chromosomal damage)
- Cross-linking of broken strands tin lead to abort of mitosis, prison cell expiry, or mutation.
DNA repair mechanisms
- Mechanisms used to right replication errors during DNA replication:
- Proofreading
- Mechanisms for unmarried-stranded DNA damage (later replication):
- Photorepair (not active in humans)
- Base of operations excision repair
- Nucleotide excision repair
- Mismatch repair
- Mechanisms for double-stranded Dna breaks (after replication):
- Homologous recombination
- Nonhomologous end joining
Proofreading and Photorepair
Proofreading
Proofreading occurs during Dna replication. DNA polymerase (the enzyme circuitous that replicates DNA):
- Has "proofreading" activity, capable of correcting replication errors
- Tin "back upwardly" when an error is detected, excise the incorrect nucleotide, and insert the correct one:
- Has 3' to five' exonuclease activity allowing this excision of the incorrect nucleotide
- Improves the fidelity of copying the Deoxyribonucleic acid 100-fold
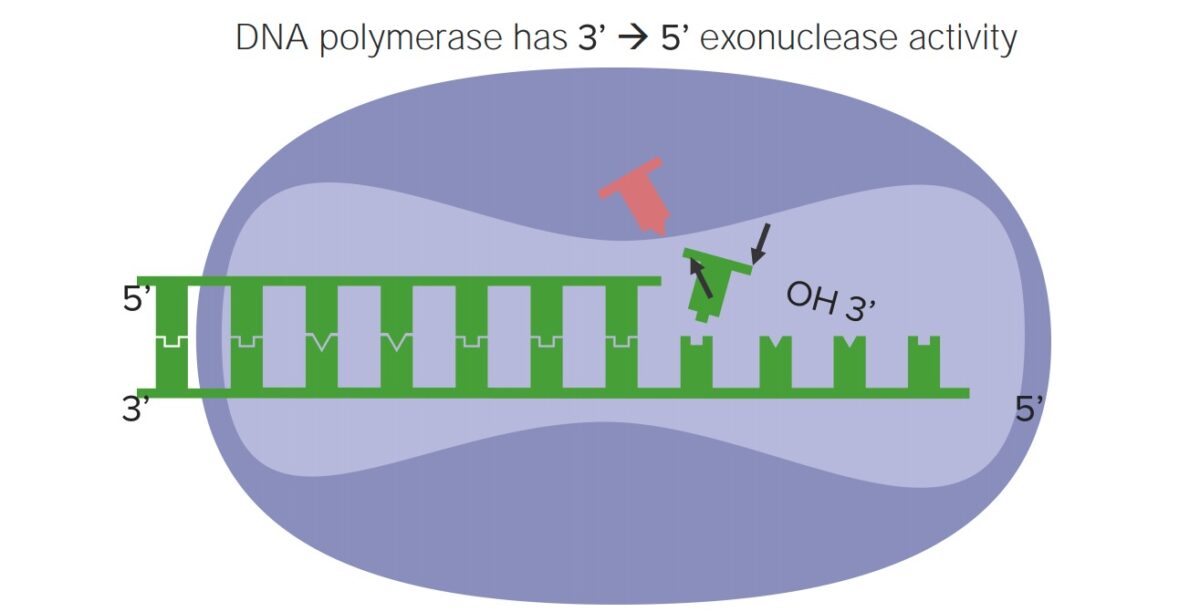
Proofreading activity of DNA polymerase
Epitome by Lecturio.Photorepair
UV light radiation causes pyrimidine (T or C) dimers to form via covalent linkages between adjacent bases, creating a conformational change ("bulge") in the DNA. These defects can be repaired via a procedure called photorepair, or photoreactivation.
- Dna photolyase is an enzyme that uses visible light energy to break the covalent bail between pyrimidines, restoring the DNA to its original state:
- Note that UV light causes the damage; visible light provides the free energy to fix the damage.
- Photorepair is an example of straight (or chemic) reversal of DNA harm (i.e., information technology does not break the phosphodiester backbone).
- Does not demand a Dna template
- Tin can repair just 1 base at a time
- This process is not active in placental mammals (such as humans).
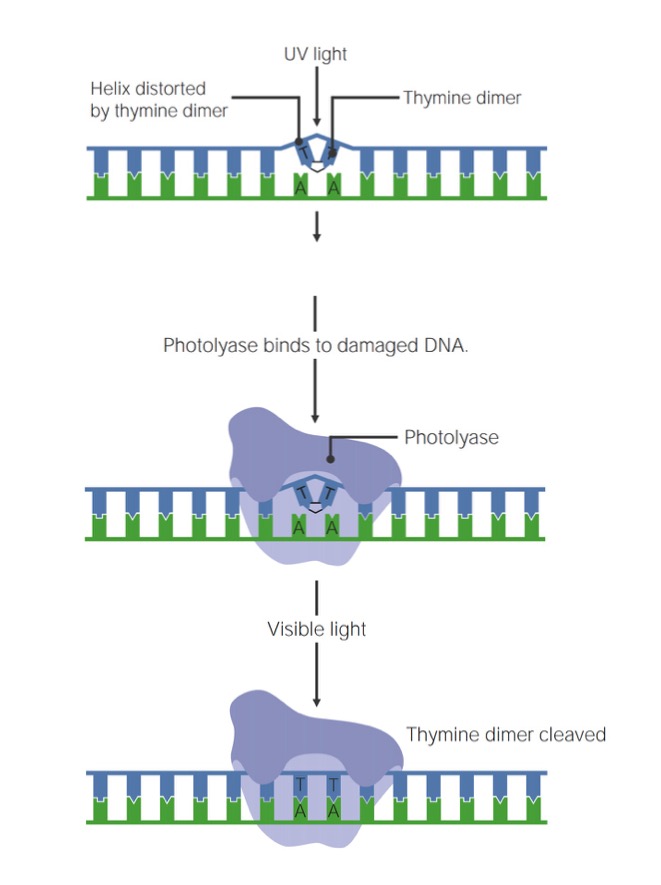
Steps of photorepair
Image by Lecturio.Single-Stranded Dna Damage Repair
The cell has three primary mechanisms to repair impairment to a unmarried strand of DNA: base excision repair, nucleotide excision repair, and mismatch repair.
Full general procedure of unmarried-stranged Dna damage repair
All 3 mechanisms follow the aforementioned full general process:
- The damaged or abnormally paired bases are identified.
- Excision enzymes excise the abnormal base, nucleotide, or a small region surrounding it.
- Deoxyribonucleic acid polymerase fills in the gap past calculation the right nucleotides.
- Deoxyribonucleic acid ligase seals the saccharide-phosphate courage with a new phosphodiester bond.
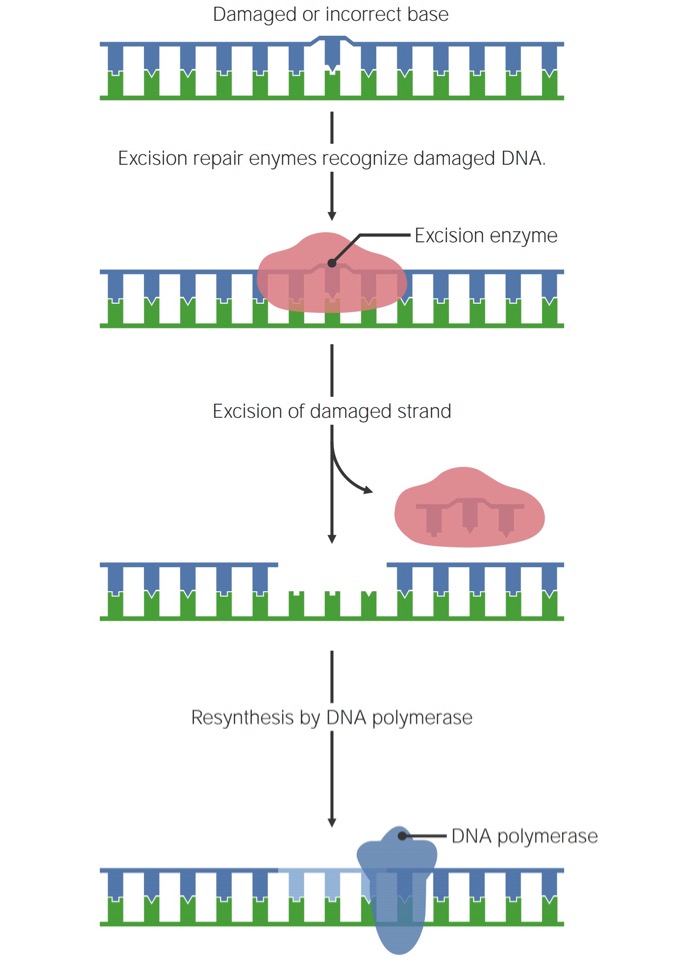
Full general mechanism of single-stranded DNA repair
Paradigm past Lecturio.Base of operations excision repair (BER)
In BER, a single damaged base is excised and replaced.
- Glycosylase enzymes:
- Cleave the bail between the damaged base and deoxyribose, removing it
- DNA glycosylases are lesion-specific.
- Cells have multiple DNA glycosylases with different specificities.
- Examples of glycosylase enzymes:
- Uracil-DNA glycosylase removes uracil from Dna strands.
- Oxoguanine glycosylase removes oxidized bases.
- Endonucleases remove the remaining saccharide-phosphate portion of the nucleotide.
- Dna polymerase removes the single abnormal nucleotide and replaces it with the correct one.
- DNA ligase seals the backbone.
- Mechanism of UV damage repair in humans
Nucelotide excision repair (NER)
- Similar process to BER, merely with larger sections of DNA excised
- The entire section of DNA (typically, approximately 12–30 base pairs) surrounding the abnormal area is removed by a complex of endonucleases.
- Deoxyribonucleic acid is resynthesized by Deoxyribonucleic acid polymerase and sealed past Dna ligase.
DNA mismatch repair (MMR)
- Corrects errors in base of operations pairing that occurred during DNA replication and were not repaired past proofreading:
- Oft caused by tautomerization (structural isomers) during replication
- Tin exist unmarried base of operations pairs or larger sections of DNA
- MMR proteins:
- Locate the mismatched nucleotides
- Recruit exonucleases to excise the mismatched bases
- Once the mismatched base is excised:
- DNA polymerase can place the correct base(s).
- DNA ligase seals the sugar-phosphate courage.
Double-Stranded DNA Damage Repair
In general, double-stranded Deoxyribonucleic acid damage is harder to repair because there is no template strand to piece of work off of. The two primary mechanisms to set double-stranded DNA breaks are homologous recombination and nonhomologous end joining.
Homologous recombination (HR)
In homologous recombination (HR), the well-nigh identical sister chromatid or homologous chromosome is used as a template:
- Mechanics and enzymes are like to the chromosomal crossing over that occurs during meiosis.
- 60 minutes requires:
- Extensive regions of homologous sequences between the 2 chromatids
- Multiple enzyme complexes, including:
- Exonucleases: begin digesting the 5' ends → generates 3' single-stranded Dna tails on the broken strands
- Recombinase enzymes: catalyze the insertion of the 3' finish into the complementary sequence of DNA on the homologous chromatid
- DNA polymerase is able to synthesize new DNA from the homologous chromatid.
- The "invading" strand may terminate up:
- Being exchanged with its homologous region on the opposite chromatid:
- This exchange is known as a Holliday junction, a second end capture, or crossing over.
- Existence displaced later it has synthesized enough Dna to "cross the gap" of its original break:
- The original 3' end is and so reconnected with its original 5' end and used as a template to fill in the gaps.
- Known as synthesis-dependent single-strand annealing (SDSA)
- Copying the entire remainder of its sister chromatid if its original five' end is lost:
- Once the invading three' cease has copied the rest of the chromatid, it releases and is used as a template strand to remake its complementary strand.
- This process is known as suspension-induced replication (BIR).
- Being exchanged with its homologous region on the opposite chromatid:
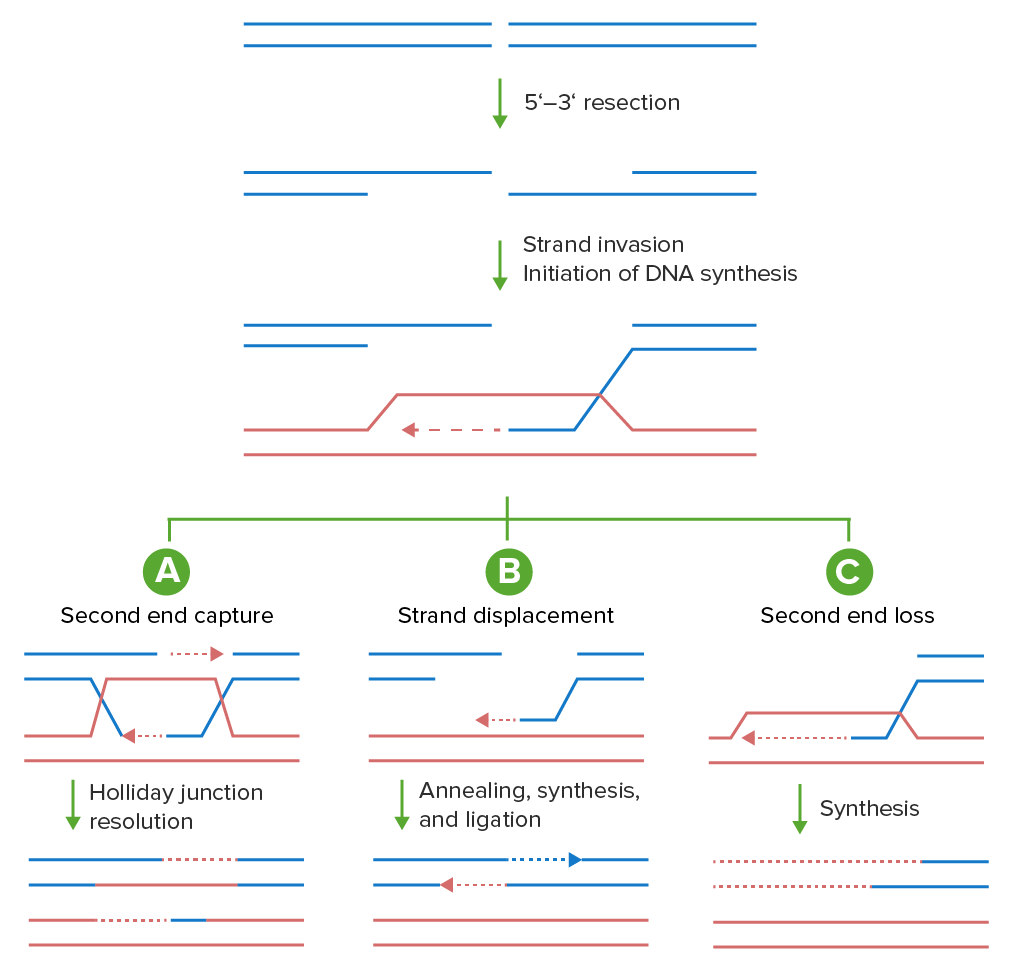
Models of homologous recombination:
Double-stranded breaks can be repaired using the homologous recombination machinery in a variety of means. The Dna ends are first processed into 3′ single-stranded DNA tails. These tails invade a homologous template (crimson), priming new DNA synthesis (dashed line). Shown are 3 possible outcomes from this invasion.
A: In canonical double-stranded interruption repair (DSBR), both the initial invading strand and the captured 2nd stop anneal to the homologous template and prime number new DNA synthesis, resulting in a double Holliday junction that tin be resolved by nucleases into a crossover or a noncrossover product (noncrossover product shown).
B: Alternatively, subsequently the single-stranded DNA tail invades the homologous template, a circular of Deoxyribonucleic acid synthesis is prepared from the three′ end (dashed red line). Synthesis-dependent strand annealing (SDSA) occurs when the invading strand, along with the newly synthesized segment, is unwound by a helicase and annealed with the other resected finish.
C: In break-induced replication (BIR), 1 finish of the DSB is lost and the remaining end invades the homologous template priming DNA synthesis to the end of the chromosome.
Nonhomologous end joining (NHEJ)
- Dna ligase Iv and a cofactor, XRCC4, directly join the cleaved ends back together.
- Relies on microhomologies (short homologous sequences) on single-stranded tails of the broken strands
- Deletions, insertions, and translocations are more common with this type of repair.
- A similar machinery is involved in V(D)J recombination to create B-cell and T-cell multifariousness.
Clinical Relevance
- Genetic disorders: When mismatched base pairs are not repaired within the gametes, they tin can create mutations that are passed on to offspring.
- Cancer: form of diseases acquired by changes in the genetic sequence and gene expression. These changes are often due to DNA mutations or harm. There are iv classes of normal regulatory genes that are frequently damaged in cancer cells: growth-promoting oncogenes, growth-inhibiting tumor suppressor genes, genes that regulate apoptosis, and genes involved in Dna repair.
- Lynch syndrome: also chosen hereditary nonpolyposis colorectal cancer (HNPCC). Lynch syndrome is caused by an autosomal dominant mutation in Dna MMR genes. Lynch syndrome is the virtually common inherited colon cancer syndrome, and it carries a significantly increased risk for endometrial cancer and other malignancies. Diagnosis is fabricated past genetic testing. Early and frequent screening for colon cancer is required, and prophylactic hysterectomy is often recommended for women beyond reproductive age.
- Skin cancers: The almost common types of skin cancers include basal cell carcinoma, squamous cell carcinoma of the peel, and melanoma. The mutations in these cancers oft arise from DNA damage due to excessive exposure to UV radiation. When overloading of BER systems occurs owing to frequent sun exposure, the take chances for skin cancers increase. This risk is about common in off-white-skinned individuals with a history of excessive sun exposure and sunburns. Definitive diagnosis is established with biopsy. Treatment relies primarily on surgical excision.
- Xeroderma pigmentosum: rare autosomal recessive disorder characterized by paint changes in the skin. The presence of xeroderma pigmentosum leads to an increased gamble for UV-induced skin and mucous membrane cancers and, occasionally, progressive neurodegeneration. This disorder is caused past ane of several mutations, which ultimately leads to dysfunctional NER. Diagnosis should be suspected in children with severe sunday sensitivity, significant freckling prior to historic period 2, and/or skin cancer prior to age 10.
References
- Friedberg, East. (2003). Dna harm and repair. Nature 421:436–440. https://doi.org/x.1038/nature01408
- Li, X., Heyer, W. D. (2008). Homologous recombination in Dna repair and Deoxyribonucleic acid damage tolerance. Cell Research 18:99–113. https://doi.org/x.1038/cr.2008.1
- Roldan-Arjona, T., Ariza, R. R., Cordoba-Canero, D. (2019). DNA base excision repair in plants: an unfolding story with familiar and novel characters. Frontiers in Establish Science. Retrieved April 23, 2022, from https://www.frontiersin.org/articles/10.3389/fpls.2019.01055/full
Source: https://www.lecturio.com/concepts/dna-repair-mechanisms/
Posted by: bettencourtmody1941.blogspot.com


0 Response to "Are Double Strand Repairs On Mcat"
Post a Comment